Beef cattle are one of the last major livestock species in the U.S. that have not moved into confinements with controlled environments. Cows spend their entire lives outside, and are raised in a variety of environments, each with a unique combination of stressors. Anecdotal and experimental data suggests that some cattle are better adapted to certain environments than others. My research attempts to identify genetic variants that control adaptation in cattle populations. We plan to use these variants to produce environment-specific breeding values that will provide producers with selection tools to identify the best animals for their region. This work also provides insight into the complex biology of adaptation in mammals.
Our ability to detect adaptive variants relies on the assumption that well-adapted cattle will go on to produce lots of offspring within their home region while poorly adapted cattle will be quickly culled from herds. We anticipate that this selection will create detectable frequency differences at alleles involved in adaptation. We identify these genomic regions using continuous and discrete definitions of climate as dependent variables in genome-wide linear mixed models. This allows us to control for the relatedness of individuals across climates and identify likely adaptive variants. We group animals into statistically-derived ecoregions (based on long-term climate data). Other work in our group is developing ecoregion-specific EPDs that will allow producers to select animals that are well-adapted to their local environment.
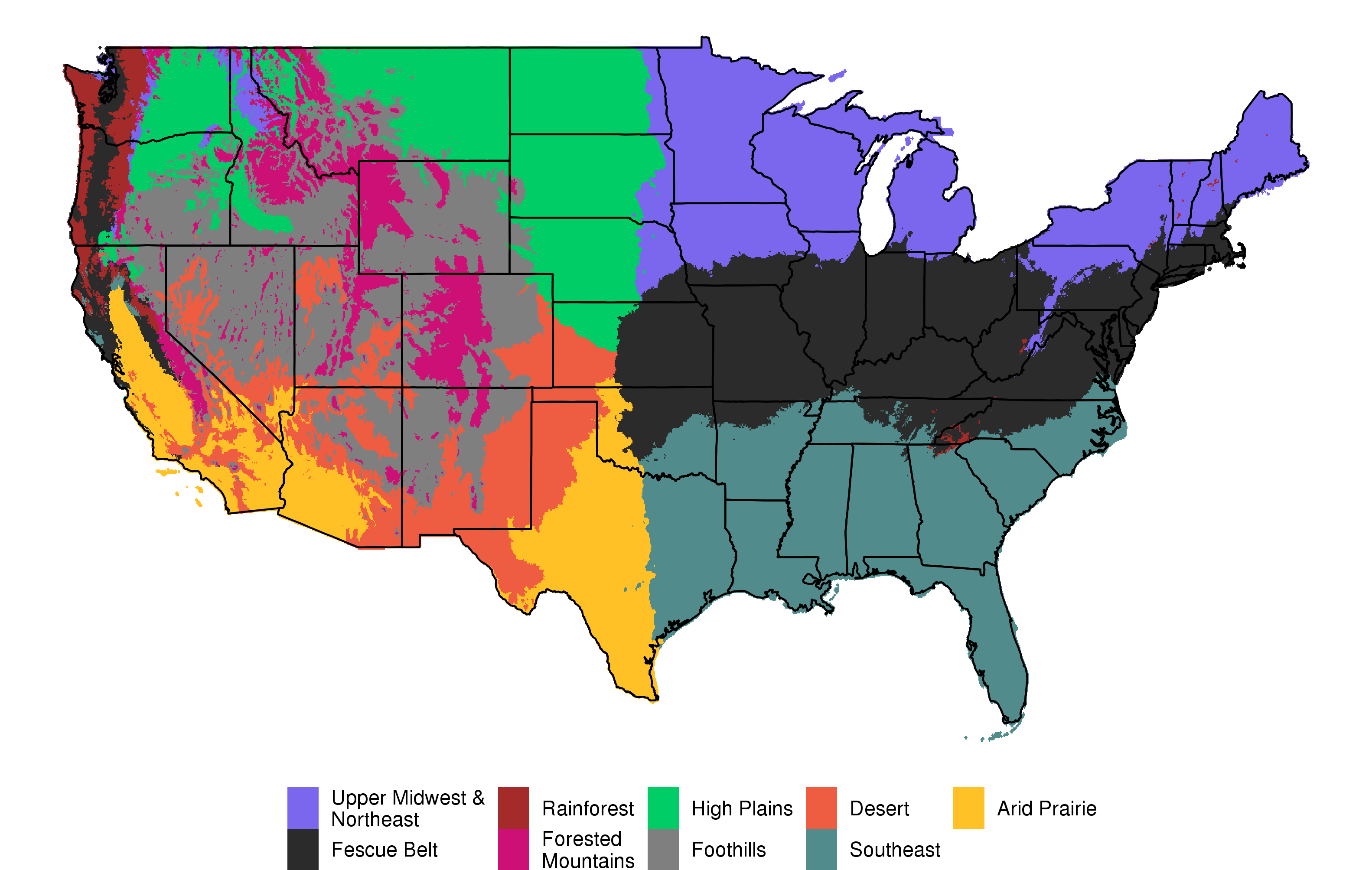
Statistically-derived ecoregions (from long-term temperature, precipitation, and elevation data)
When applied to U.S. cattle populations, we identify dozens of genomic regions in each breed associated with environment. These areas are enriched for genes that play roles in neural development and signalling, as well as in the secretion of hormones involved in vasoconstriction and vasodilation. We suspect that these adaptations allow animals to adapt to a number of stressors like heat, cold, and toxic fescue. When we search for loci undergoing ecoregion-specific selection (changing in frequency in one region, but not the others), it appears that local adaptation is being eroded through the use of artificial insemination as opposed to being positively selected for.
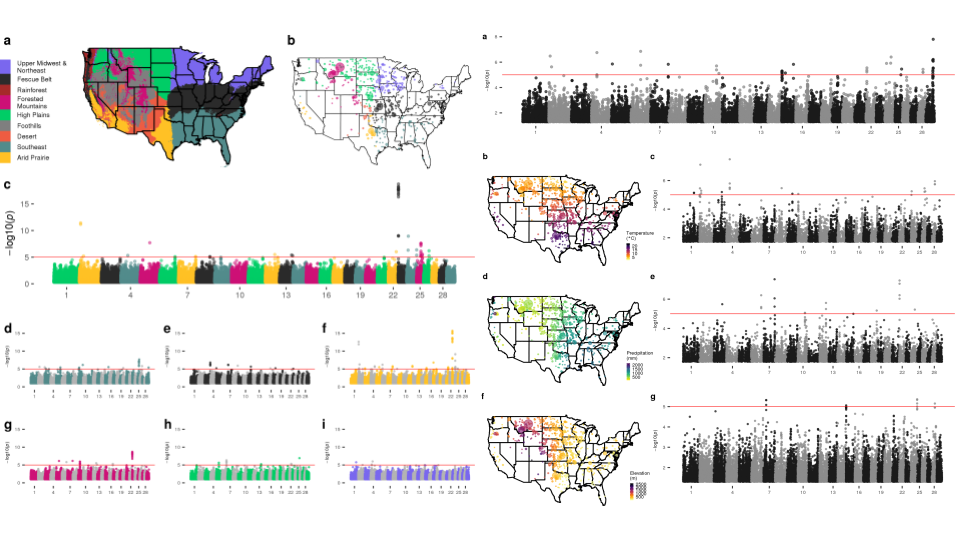
We fit discrete ecoregions and continuous environmental variables as dependent variables in GWAS models to map local adaptation and identify dozens of genomic regions.
For a more in depth look at this work, read our preprint!
